Barking up the right tree: an approach to search over molecule synthesis DAGs
Super Short Description
- Paper Link
- This work is on generation of molecules using a generative model. Novelty of the paper is that it also outputs the molecule synthesis DAG: A step by step sequence of how the generated molecule can be constructed. Another interesting work done in the paper is that a specific training procedure ensures that molecules with targeted properties are generated.
An Overview of Methodology
Starting from a selected set of building blocks (simple chemical compounds), the molecule is built one step at a time. Paper defines different action types. Sequentially selecting appropriate actions, each belonging to one of those action types leads to the synthesis DAG construction. Qualitatively, actions include the choice of reactants, the number of reactants in a reaction and the number of reactions. Note that there are two sets for the choice of reactants: original building blocks and the intermediate compounds formed till that point in time. These actions are predicted from their respective action-type neural networks. For predicting the DAG, the paper represents DAG as a sequence of actions and it predicts one action at a time.
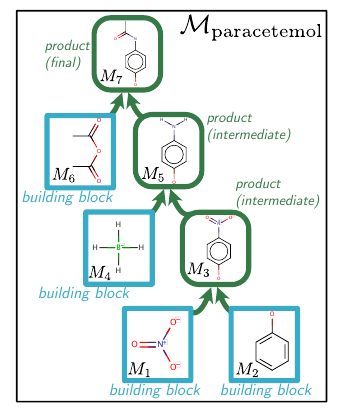

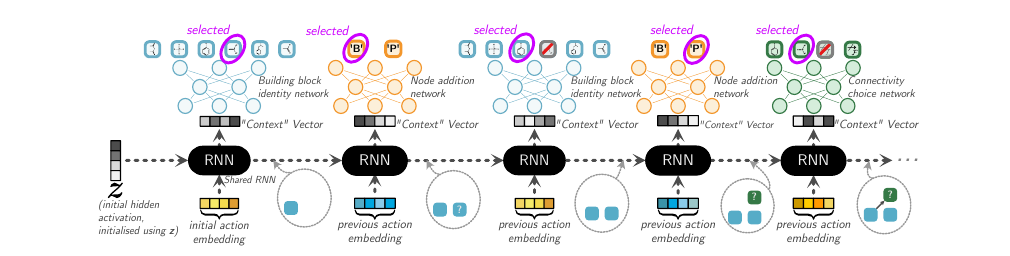
Action Types
There are three types of actions in the construction of the serialized DAG.
- (A1) Node Addition: It determines the type of reactant in a reaction. It can be either an intermediate product (denoted by ‘P’) or one of the building blocks (denoted by ‘B’).
- (A2) Building block molecular identity: Once the type of reactant is decided, this action decides which specific chemical compound belonging to the type is to be used. In the serialized DAG, A2 is constrained to immediately follow A1.
- (A3) Connectivity Choice: This decides what reactants are to be used for the current reaction. It links existing nodes (‘P’ type or ‘B’ type) to the freshly created product node. It also decides whether the product formed is intermediate or final.
Definitions:
- Final Molecule is \(M\)
- Synthesis DAG \(M\) is composed of a sequence of actions \(V^i\), \(M=[V^1,V^2,..V^L]\). Also, action type of \(V^i\) is \(A^i, A^i\in\{A1,A2,A3\}\).
- \(V_{<l}\) denotes sequence of actions taken before \(V_l\).
Probablistic Model Based Architecture
They define the probablity distribution over the actions as follows.

They use a common RNN to model each term shown above. Specifically, for every action type, there is a neural network. This neural network takes as input the output of the RNN. This models the conditionality on previous actions. However, the hidden state of the RNN does not have the explicit information about what the last predicted action was. So, the input to the RNN is the action embedding for the last action which was taken. If the action involved a molecule, (like A2,A3) then GNN was used to get the embedding for the molecule which gets used as the action embedding. Otherwise (action type A1), the embedding vector is learnt for these abstract actions. In this setup, the initial hidden state of the RNN has significance since it is that which has the potential to guide the construction towards a specific subdomain of molecules.
Two Model Variants
In the first variant, they create DoG-AE, an autoencoder based model with decoder being the above described model. They create an encoder using graph neural network. After randomly tweaking the latent code obtained from the output of the trained encoder, they show that they are able to generate similar molecules.
In the second variant DoG-Gen, they fix the latent code (which becomes the latent vector for the first step in RNN) to zero. They fine-tune the weights of the network so as to allow generation of molecules with certain properties.
